From Code to Context: Interpreting 21 Million Rows of CRISPR Data
In a recent post, I detailed the “how” of unpivoting 21 million rows of genomic data using DuckDB. Since then, the project has matured significantly. I’ve implemented Dagster for orchestration and moved the compute to a Spark-on-Delta Lakehouse architecture.
In data engineering, architectural complexity is often a tax, and sometimes you do not know if it will pay off until you are further along the project. I probably spent more time on just JVM memory issues than I did on the whole previous post. However, once I had the Spark job running and managed by Dagster, measuring and validating the data became straightforward.
Understanding the Bioinformatics Benchmarks
For those coming from a pure data engineering background, genomic “Effect Scores” can be opaque. When scientists use CRISPR, they are essentially “deleting” a gene from a cell’s DNA to see if the cell survives.
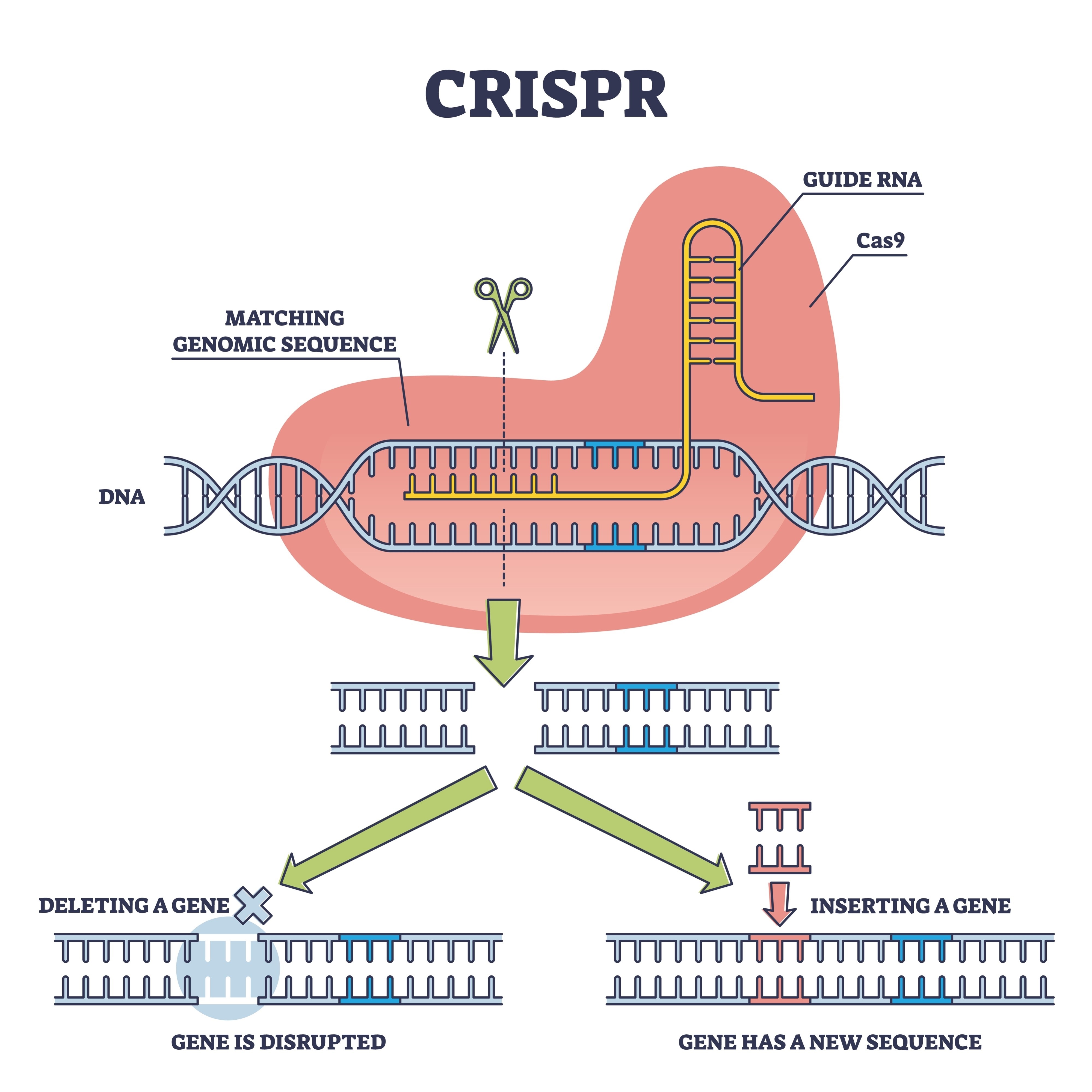
The result is a Dependency Score:
- 0.0 (Neutral): Deleting the gene had no impact.
- -1.0 (Essential): This is the “kill switch” threshold; the gene is necessary for life.
To ensure my new architecture hadn’t corrupted the data during the transition to a relational “Gold” layer, I used Spark to aggregate the scores for all 17,000+ genes across 1,100+ cancer cell lines.
The Results: A Biological “Sanity Check”
The results from the pipeline were striking:
- RPL15: -5.69
- SNRPD3: -5.47
- RAN: -4.23
If you aren’t a biologist, these look like random strings. But to a Bioinformatician, they are Common Essentials—the “housekeeping” genes.
RPL15 builds ribosomes (protein factories), and SNRPD3 handles RNA processing. Because these scores are so extreme (well beyond -1.0), we know these genes are “pan-essential.” Seeing these known biological truths at the top of the list validates the entire engineering stack. It proves the unpivot logic is mathematically sound.
The State of the Industry: Seeking Selective Vulnerabilities
In modern Bioinformatics, identifying SNRPD3 isn’t the end goal—it’s the baseline. Because these genes are essential to all cells, they make poor drug targets; you can’t kill a cancer cell by destroying machinery that healthy cells also need.
The real “Gold” lies in finding Selective Essentials: genes that are a -1.0 in Breast Cancer but 0.0 in healthy tissue.
The Path Forward: Data Enrichment
The move to a searchable Lakehouse wasn’t about raw speed; it was about turning a static file into a foundational platform. With the data now indexed in Delta tables and orchestrated by Dagster, we can begin Data Enrichment:
- Clinical Correlation: Joining “Achilles’ heels” with patient mutation data from the TCGA (The Cancer Genome Atlas).
- Druggability: Cross-referencing targets with the Drug Gene Interaction Database (DGIdb).
- Selective Search: Filtering for vulnerabilities unique to specific cancer subtypes.
By building this infrastructure, we’ve moved beyond a one-off script and created a relational engine ready for the next level of genomic discovery.