Architecting a High-Performance Research Platform for CRISPR Dependency Maps
The Challenge: Normalizing Multi-Dimensional Matrices
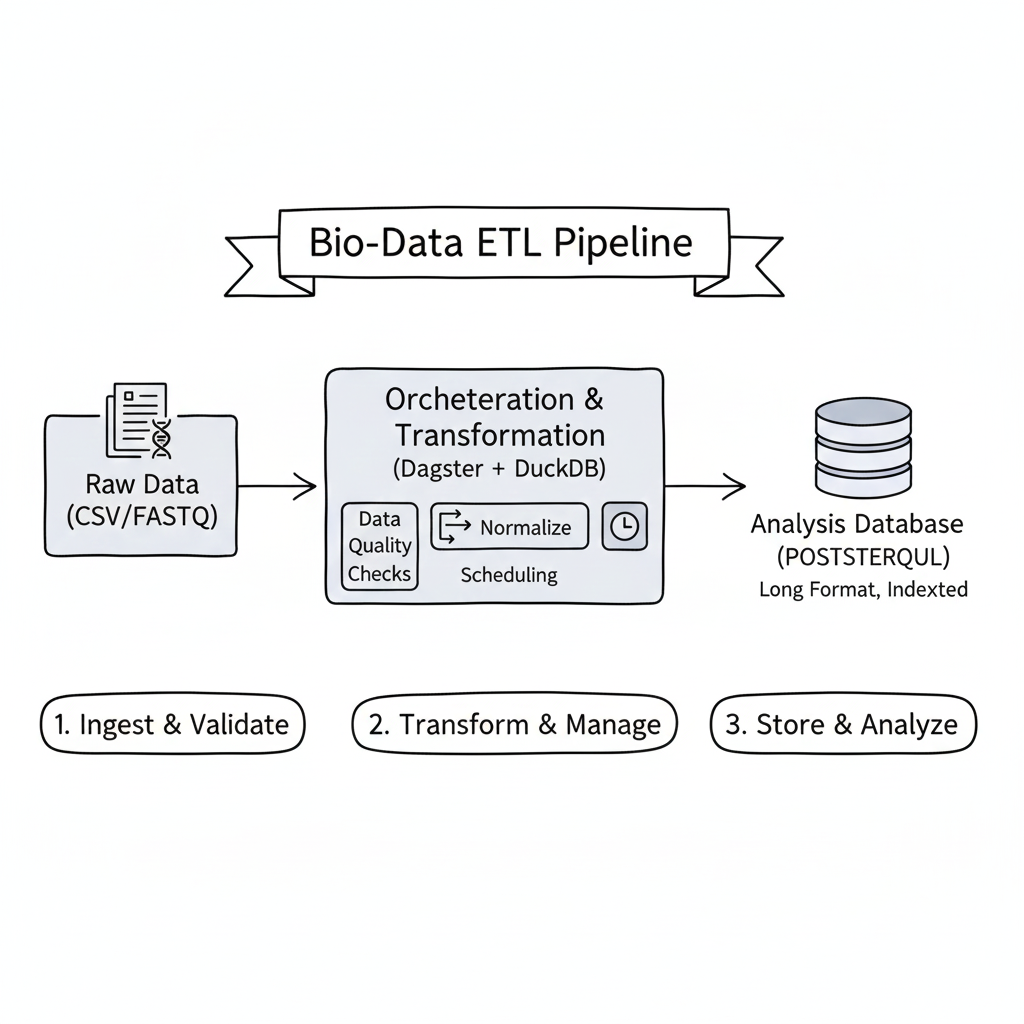
The DepMap (Cancer Dependency Map) dataset is a standard resource in genomics, but like many large-scale research assets, it is delivered as a “Wide” matrix. In this case, the raw CRISPR data contains 17,000 columns representing individual gene effects. While this structure is common for initial data collection, it is a non-starter for relational analysis. Querying a specific gene across thousands of cell lines is inefficient when the data is structured this way. To make this information actually usable for downstream research—adhering to FAIR (Findable, Accessible, Interoperable, Reusable) principles—the first step is normalizing that matrix into a “Long” format. Once the initial transform is complete, we have a 21-million-row table—still large, but far more manageable.
Building Production-Grade Infrastructure
Building an environment to manage this type of data requires moving beyond basic scripts into production-grade infrastructure. I am using a Fedora workstation running in my local environment; while this could just as easily run in a cloud VM or bucket, here I am taking advantage of convenience. The project is fully transferable to a production environment by running any type of S3-compatible container.
- Compute: DuckDB is the practical choice for this transformation; its vectorized execution handles “unpivoting” high-dimensional matrices without the memory overhead that typically crashes Pandas or standard SQL.
- Persistence: To ensure the database remains persistent and auto-restarting, I used Podman Quadlets to manage the PostgreSQL instance as a systemd service. This effectively turns a local container into a reliable piece of Infrastructure-as-Code.
- Orchestration: The final requirement for a biotech pipeline is observability. Because DepMap data updates on a semi-annual schedule, it is easy for pipelines to break quietly due to schema drift or corrupted downloads. I used Dagster for orchestration to ensure the system flags errors at the ingestion stage before they hit the research database. This moves the project from a “one-off” load to a managed data lifecycle.
Persistent Infrastructure: The Quadlet Configuration
To move beyond a transient container, I utilized Podman Quadlets to manage the PostgreSQL instance. By defining the container in a .container file, Fedora’s systemd handles the lifecycle—starting the database at boot and ensuring it auto-restarts if it fails.
First, I defined the container configuration:
# ~/.config/containers/systemd/crispr-db.container
[Container]
Image=docker.io/library/postgres:latest
Environment=POSTGRES_PASSWORD=YOUR_PASSWORD_HERE
Environment=POSTGRES_DB=crispr_db
PublishPort=5432:5432
Volume=postgres_data:/var/lib/postgresql/data
[Service]
Restart=always
[Install]
WantedBy=default.targetThen, I used systemctl to load the Quadlet and start the service:
# Reload systemd to recognize the new Quadlet
systemctl --user daemon-reload
# Start and enable the database service
systemctl --user enable --now crispr-db.service
# Verify the container is running
podman psThis setup provides a persistent “Silver” layer that feels like a production cloud instance but runs entirely on local hardware.
The Implementation: DuckDB to Postgres Stream
The actual execution was handled via DuckDB’s Postgres scanner. By unpivoting all 17,000 columns and streaming them directly into the containerized database, the final table was populated in under 4 minutes.
-- Normalizing and Loading 21M rows in a single pass
INSTALL postgres;
LOAD postgres;
-- Connecting to the Podman-backed instance
ATTACH 'host=localhost user=postgres password=YOUR_PASSWORD_HERE dbname=crispr_db' AS pg (TYPE POSTGRES);
-- Unpivot 17k columns into a "Long" format for indexing
CREATE TABLE pg.gene_effects AS
SELECT
"column00000" AS model_id,
gene_symbol,
dependency_score
FROM (
UNPIVOT (SELECT * FROM read_csv_auto('CRISPRGeneEffect.csv'))
ON COLUMNS(* EXCLUDE "column00000")
INTO NAME gene_symbol VALUE dependency_score
);Strategic Reflection: Traceability and Integrity
Ultimately, this project is about establishing Data Provenance, but the primary importance of this system lies in its computational efficiency for therapeutic discovery. In a regulated GxP environment, the process behind the data is as important as the data itself. A researcher can now query a genetic target in milliseconds, knowing that every row is backed by a versioned, observable audit trail. Moving from a “Wide” CSV silo to a high-performance platform like this is what turns raw research data into a strategic asset for drug discovery.
Expanding the Infrastructure: Next Steps
From here, we can build on this infrastructure to move beyond simple storage and into Cross-Domain Discovery:
- Multi-Omics Integration: By joining this CRISPR data with the GWAS (Genome-Wide Association Study) catalog, we can cross-reference genetic variants found in patient populations with the dependency scores found in the lab. This helps prioritize “High-Confidence” targets for drug development.
- CDISC/SDTM Compliance: We can extend the pipeline to map this “Silver” layer data into SDTM (Study Data Tabulation Model) domains. This demonstrates the ability to transform raw research hits into regulatory-ready datasets for clinical submissions.
- Target Identification Dashboards: With the data now indexed in Postgres, we can build a Streamlit or React interface that allows scientists to visualize gene “essentiality” across different cancer lineages (e.g., Lung vs. Breast) in real-time.